Contents
% Risk, Bayes Risk % Bias-Variance decomposition % Overfitting/Underfitting % % (c) Faïcel Chamroukhi %%%%%%%%%%%%%%%%%%%%%%%%%%%% clear; close all; clc warning off; set(0,'defaultaxesfontsize',16); %plot option
Data generating process
% Consider the following arbitrary target function f = @(x) 10 + 5*x.^2.*sin(2*pi*x); % the true function
plot the true function
fplot(@(x) f(x), [0, 1],'k--', 'linewidth', 1.5); xlabel('x');ylabel('y') legend('True f(x)','Location','northeast')

Consider the following data generating process:  ; for
; for 
n = 20; % x_i's fixed or random x = linspace(0,1, n)'; % e: independent Gaussian noise e parameters \mu_e and \sigma_e: e \sim \mathcal{N}(\mu_e, \sigma_e^2) mu_e = 0; sigma_e = 1; e = normrnd(mu_e, sigma_e, n, 1); % an example of one sample y = f(x) + e;
An illustrating example: plot the illustrating example
figure % plot the true function fplot(@(x) f(x), [0, 1],'k--', 'linewidth', 1.5); xlabel('x');ylabel('y') legend('True f(x)','Location','northeast') % together with the observed noisy pairs (x_i, y_i) hold on; plot(x,y,'ko', 'MarkerSize',12) legend('True f(x) ','Observations (x_i,y_i)') pause

Polynomial regression
% Consider a ploynomial regression model of order p p = 4; % the fitted OLS function %X = RegDesign(x, p); beta_OLS = polyfit(x, y, p); %(X'*X)\X'*y; %inv(X'*X)*X'*y; f_hat = @(x) polyval(beta_OLS, x); % plot the fitted function hold on, fplot(f_hat, [0, 1],'r-', 'linewidth', 1.5); ylim([mean(y)-2*std(y), mean(y)+3*std(y)]) legend('True f(x) ','Observations (x_i,y_i)',['Fitted f(x): Polynomial of order p=',int2str(p)]) pause

Consider ploynomial regression functions, with increasing polynomial degree  , from 0 to
, from 0 to 
pmax = 14;
Examples of # polynomial fits
plot_examples = 0; if plot_examples
for p=0:pmax
compute the OLS Estimator
beta_OLS = polyfit(x, y, p);
% Identical to:
%
% X = RegDesign(x, p);
% beta_OLS = (X'*X)\X'*y; %inv(X'*X)*X'*y;
compute the fitted values of the OLS function
f_hat = @(x) polyval(beta_OLS, x);
plot the true function, together with the observed noisy pairs (x_i, y_i) and the fitted OLS function:
figure
% plot the true function
fplot(@(x) f(x), [0, 1],'k--', 'linewidth', 1); xlabel('x');ylabel('y'); legend('True f(x)','Location','northeast')
% plot the observed noisy pairs (x_i, y_i)
hold on, plot(x,y,'ko', 'MarkerSize',12)
legend('True f(x) ','Observations (x_i,y_i)')
% plot the fitted OLS function
hold on, fplot(f_hat, [0, 1],'r-', 'linewidth', 1.5); ylim([mean(y)-2*std(y), mean(y)+2*std(y)])
legend('True f(x) ','Observations (x_i,y_i)',['Fitted f(x): Polynomial of order p=',int2str(p)])
pause







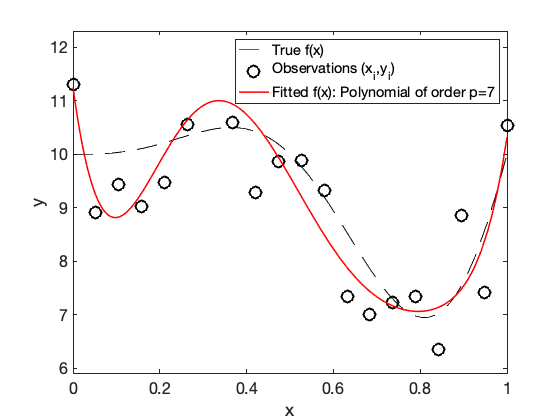







end % end pause
Learn and evaluate different prediction functions
Consider N sample replicates to average the results
N = 100; % Compute the traning and testing erros, the bias and the variance for each prediction function Training_error = zeros(N, pmax+1); % The training error for each sample Testing_error = zeros(N, pmax+1);% The prediction error for each sample % for later computing of the biases and the variances Hx = zeros(n, pmax+1, N); % Matrices to store all the prediction function values Fx = zeros(n, pmax+1, N); % Matrices to store all the true function value
Let's consider for now a fixed design (the x's are a uniform grid in [0, 1]
x = linspace(0, 1, n)'; % for training xtest = linspace(1/n, 1, n)'; % for testing
Draw N samples to average the results
for sample=1:N
draw a training sample
y = f(x) + normrnd(mu_e, sigma_e, n, 1);
draw a testing sample
ytest = f(xtest) + normrnd(mu_e, sigma_e, length(xtest), 1);
for p=0:pmax
train the model (OLS):
beta_OLS = polyfit(x, y, p);
fitted OLS function
f_hat = @(x) polyval(beta_OLS, x);
Training Error (under the square loss)
Training_error(sample,p+1) = 1/length(x) * sum((y - f_hat(x)).^2);
Testing Error (under the square loss)
Testing_error(sample,p+1) = 1/length(xtest) * sum((ytest - f_hat(xtest)).^2);
Store the fitted h's and the trure values of the f's for later calculation of the biases and the variances
Fx(:, p+1, sample) = f(xtest);
Hx(:, p+1, sample) = f_hat(xtest);
end
end
Plot the results
boxPlot the training error and the prediction error
figure, boxplot(Training_error), ylim([0, 7]) xlabel('Model complexity ($p$)'); ylabel('Training Error (square loss)') figure, boxplot(Testing_error), ylim([0, 7]) xlabel('Model complexity ($p$)'); ylabel('Prediction Error (square loss)') % % Error and Prediction erros plotted together % figure,subplot(121) % boxplot(Training_error) % ylim([0, 7]) % xlabel('Model complexity ($p$)'); ylabel('Training Error (square loss)') % subplot(122), boxplot(Testing_error) % xlabel('Model complexity ($p$)'); ylabel('Prediction Error (square loss)') % ylim([0, 7]) % set(gcf,'Position',[100 500 1500 500]) pause


compute the mean errors
mean_training_error = mean(Training_error,1); mean_testing_error = mean(Testing_error,1);
plot the mean errors
figure, plot(0:pmax, mean_training_error,'k', 'linewidth', 1.5) hold, plot(0:pmax, mean_testing_error ,'r', 'linewidth', 1.5) xlim([0, pmax]); ylim([0, 7]) xticks(0:pmax); xlabel('Model complexity ($p$)') ylabel('Error (square loss)') legend('Mean Training Error','Mean Test Error','Location','NorthEast') pause
Current plot held

plot the error bars
figure, errorbar(0:pmax, mean_training_error, std(Training_error,1), 'k') hold on, errorbar(0:pmax, mean_testing_error, std(Testing_error,1),'r') xticks(0:pmax); xlim([0, pmax]); ylim([0, 7]) xlabel('Model complexity ($p$)') ylabel('Error (square loss)') legend('Training Error','Prediction Error','Location','NorthEast') pause

Another plot type with shaded regions (using the boundedline function)
figure, [hl,hp] = boundedline(0:pmax, mean_testing_error, std(Testing_error,1) , 'r-','linewidth',1, 'transparency', 0.1); hold on, plot(0:pmax, mean_testing_error, 'k', 'linewidth', 2); ho = outlinebounds(hl,hp); set(ho, 'linestyle', '--', 'color', 'r');%, 'marker', '.');set(ho, 'linestyle', '-', 'color', 'r', 'marker', '.'); xticks(0:pmax); xlim([0, pmax]); ylim([0, 7]) box on xticks(0:pmax); xlabel('Model complexity ($p$)','interpreter','latex'); ylabel('Estimate of Risk $R = {\bf E}[(Y - h(X;\theta_p))^2]$','interpreter','latex'); %legend([h1,h2,hl,hp],'\beta_0 + \beta_1 x','(x_i,y_i)\sim p_\theta','OLS Line',['CI\_',num2str((1-alpha)*100),'%','']) % %%plot(0:pmax, mean_training_error, 'k-','linewidth',2); %%hold on, plot(0:pmax, mean_training_error - std(Training_error,1), 'k--', 0:pmax, mean_training_error + std(Training_error,1),'k--') %hold on, plot(0:pmax, mean_testing_error, 'r-','linewidth',2); %hold on, plot(0:pmax, mean_testing_error - std(Testing_error,1), 'r--', 0:pmax, mean_testing_error + std(Testing_error,1),'r--') pause

Bias-Variance Decomposition
Compute the Biases and the Variances
E_h = mean(Hx, 3); % mean over the N sample replicates B2 = zeros(n,pmax+1,N); V = zeros(n,pmax+1,N); for s=1:N B2(:,:,s) = (E_h - Fx(:,:,s)).^2; V(:,:,s) = (E_h - Hx(:,:,s)).^2; end Bias2 = mean(B2,3); % all the means^2 Variance = mean(V,3); % all the variances % Bias2 = (mean(Bias2, 1)); % Bias^2 Variance = mean(Variance, 1); % Variance % Plot the Biases and the Variances figure, plot(0:pmax, Bias2,'k', 'linewidth', 1.5); xlim([0, pmax]);xticks(0:pmax) hold on, plot(0:pmax, Variance,'r', 'linewidth', 1.5); xlim([0, pmax]);xticks(0:pmax) ylim([0, 7]) xlabel('Model complexity ($p$)') ylabel('Bias - Variance') legend('Bias^2','Variance','Location','NorthEast') pause
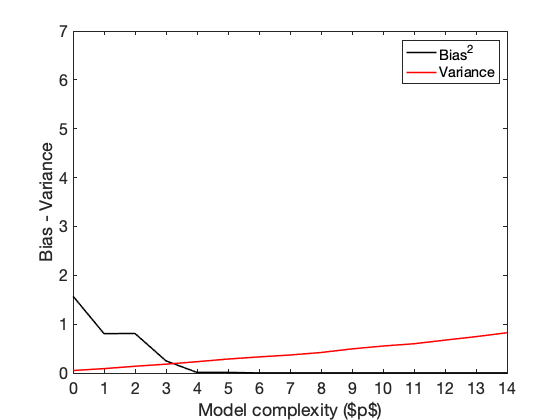
Plot all the error types together
figure, plot(0:pmax, Bias2,'k', 'linewidth', 1.5) hold on, plot(0:pmax, Variance,'r', 'linewidth', 1.5) hold on, plot(0:pmax, Bias2 + Variance + sigma_e^2,'b', 'linewidth', 1.5) hold on, plot(0:pmax, mean_testing_error ,'c', 'linewidth', 1.5) xlim([0, pmax]); xticks(0:pmax); ylim([0, 7]) xlabel('Model complexity ($p$)') ylabel('Errors | Bias-Variance') legend('Bias^2','Variance', 'Bias^2 + Variance + Bayes Error','Prediction Error','Location','NorthEast') pause

plot of bayes and prediction errors
% Bayes Error (under the square loss)
Bayes_error = sigma_e^2;
figure, plot(0:pmax, Bias2,'k', 'linewidth', 1.5) hold on, plot(0:pmax, Variance,'r', 'linewidth', 1.5) hold on, plot(0:pmax, Bias2 + Variance + sigma_e^2,'b', 'linewidth', 1.5) hold on, plot(0:pmax, mean_testing_error ,'c', 'linewidth', 1.5) hold on, plot([0 pmax], [Bayes_error Bayes_error] ,'k--', 'linewidth', 1.5) xlim([0, pmax]); xticks(0:pmax); ylim([0, 7]) xlabel('Model complexity ($p$)') ylabel('Errors | Bias-Variance') legend('Bias^2','Variance', 'Bias^2 + Variance + Bayes Error','Prediction Error', 'Bayes Error','Location','NorthEast') pause

Plot the prediction error and the Bayes error
figure, plot([0 pmax], [Bayes_error Bayes_error] ,'k--', 'linewidth', 1.5) hold on, plot(0:pmax, mean_testing_error ,'r', 'linewidth', 1.5) xlim([0, pmax]); xticks(0:pmax); ylim([0, 7]) xlabel('Model complexity ($p$)') ylabel('Errors') legend('Bayes Error','Prediction Error','Location','NorthEast')
